This document has hidden notes
Github Repository
research-experiments/ego-cells-markdown
Introduction
This experiment explores a specific use case of 📝 Ego Cells: using them to parse a markdown text buffer into an abstract representation with the possibility to use the cells' final state to generate valid HTML.
The original purpose was to test ego cells with a real-world problem. But as I worked on this experiment I realized that the value was found in exploring and learning about the nature of cellular automata and the almost infinite ways in which they can be used to represent complex systems in spacetime.
This experiment has also served as a way to explore parallel computing — working with OpenCL and implementing simple rules per cell allows me to think in a different space that I'm not used to compared to that of modern software development.
Theoretical Assumption
For as simple as markdown may seem, it requires quite a few building blocks that I didn't take into account when I was initially thinking about this first experiment. Trying to find the neighboring rules needed to create this parsing cell tissue lead me to discovering that there is an unknown number of ways to describe markdown — it's not just characters in a grid and a high-level model.
For example, I thought there had to be a purely abstract representation of truth. In this case truth being markdown syntax rules. What I realized is that truth cannot exist purely, instead, one rather translates the learned model from one space to another. I'm calling this process "Model Translation", the act of reproducing a learned system in a different modeling substrate.
Another naive assumption was that I could implement a genetic algorithm that could find the best neighboring rules to represent markdown. The problem is that due to the large number of ways in which ego cells can connect, it would take too much time to arrive to good neighboring rules. It would also require a large set of markdown samples so that the cells are able to parse unseen markdown texts effectively.
In general, there were many ideas that didn't work as I expected. But what was initially a rough concept called "ego cells" turned into a more robust building block I can use for future work. Next, I'll talk in more detail about the results and discoveries I made through this experiment.
Ego cells in detail
In the beginning, I was struggling to find a way to write the neighboring rules. I thought that each layer of cells would represent an abstraction layer.
Layer 1: raw characters
Layer 2: special characters (#, *, abc, 123)
Layer 3: tokens (heading 1 start, heading content, etc)
Layer 4: lines (heading 1, code block, paragraph, list)But following this approach failed and produced weird behavior like in the screenshot below.
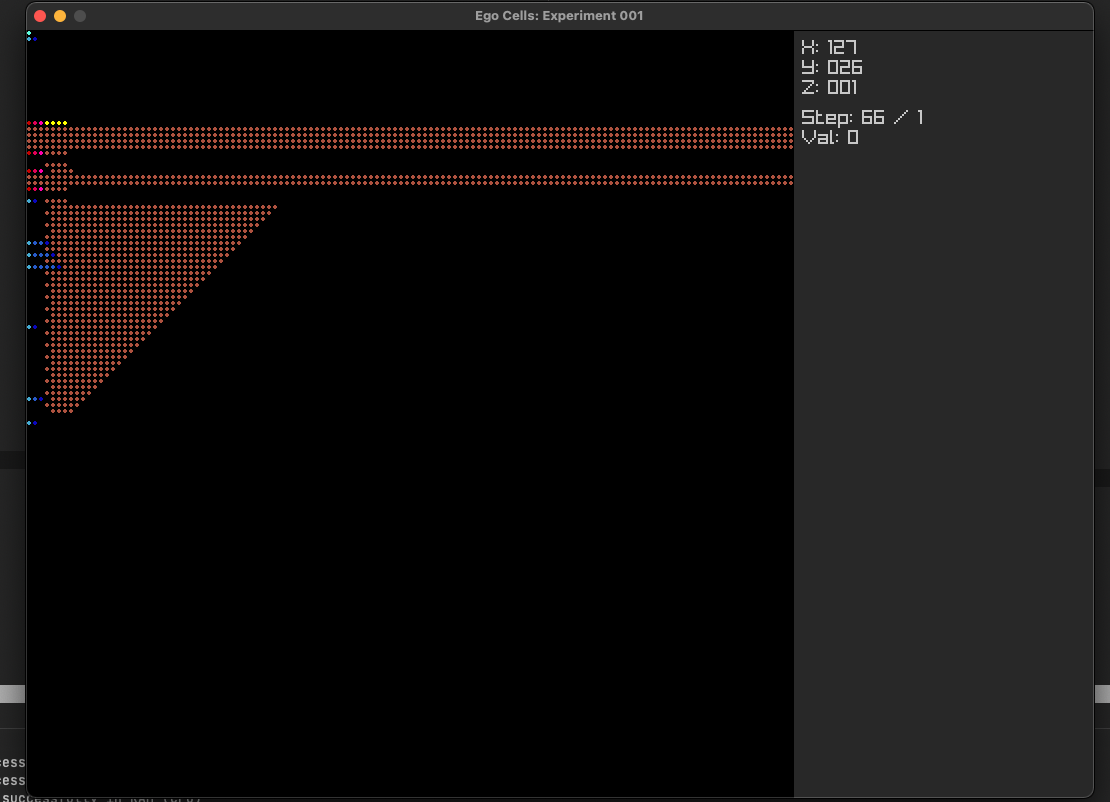
After realizing this wasn't going to work as I needed it to, I decided to go back to the basics and think about the fundamentals of markdown and text.
At this time I started reading "A new kind of science" by Stephen Wolfram. In chapter 3, he talks about the notion of simple programs. He explains that you don't need complex programs to produce complex behavior and shows multiple examples.
I turned my mindset around this and decided to throw away the abstraction approach. Instead, I focused on getting simple neighboring rules that produced the small behavior I needed. This changed everything, now I was able to rely on the small programs to know the boundaries that each cell needed to determine its state. Everything started to make sense.
Two modes of parsing
Reading is sequential and requires orientation. For example, in english we read left-to-right and top-to-bottom. I had completely ignored this fact and was struggling to find a way to parse markdown in a fully parallel way. I kept thinking that we humans can take a glimpse at a markdown text file and get a quick map of where are the headings, paragraphs, etc. I supposed this is how it should be done with ego cells, and I wasn't wrong, I simply was ignoring the sequential nature and orientation information required to properly identify different block types.
I then remembered the concept of slow-vs-fast thinking. Slow thinking requires to go through each word and follow through. Fast thinking is less precise but may provide the information we're looking for like finding a heading quick. The problem with fast thinking is that the information could be mistaken.
Take for instance the case where there is a code block with a large bash script.
...
# This looks like a heading!!
...
Let's also say that you don't speak english and you don't know what code is, and the only thing you know is markdown syntax. You could easily mistake this by a heading. This is essentially what I was struggling with on a high-level.
I thought I could solve this problem with a memory of sorts that would allow a cell to know about it's overall context — if it's within a code block or not. But it seemed overkill for this problem. So what I ended up doing was to use a layer of cells where each cell had the simple program of detecting the border and corners to decide whether or not it's part of a code block or regular text such as headings and paragraphs.
Using the example above, the cells in that bash comment are now able to know if they are a heading or just code content.
Programs exist
Using simple multi-layer programs
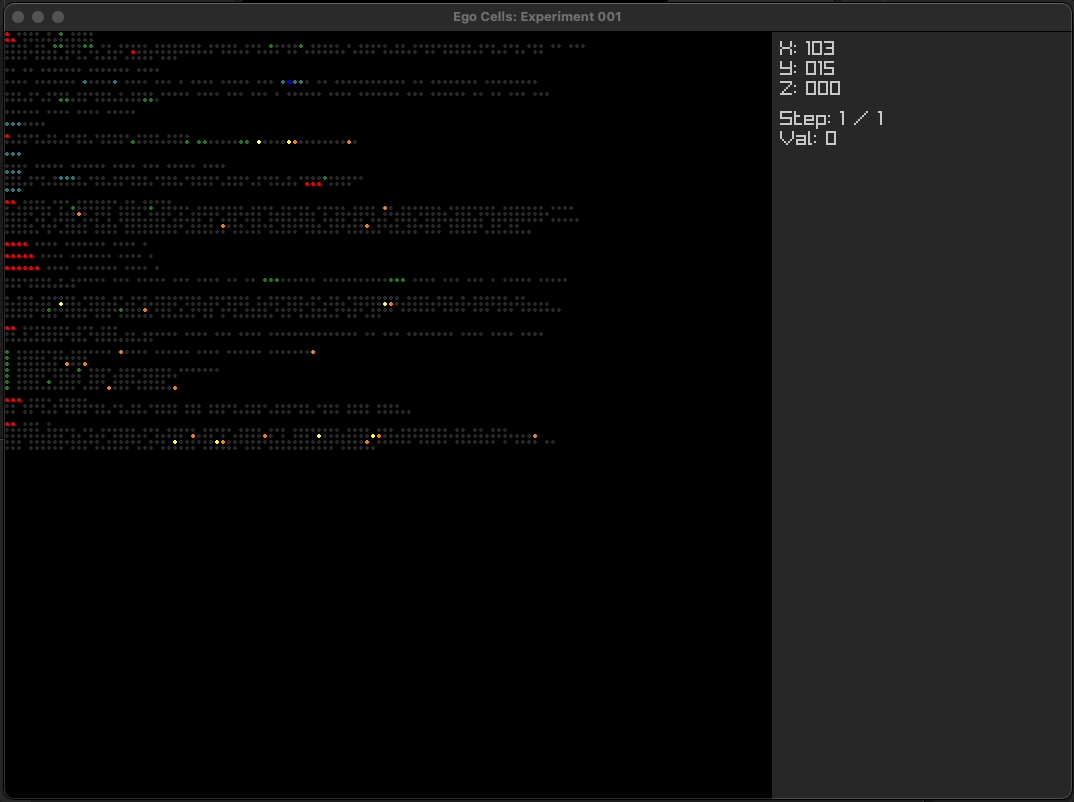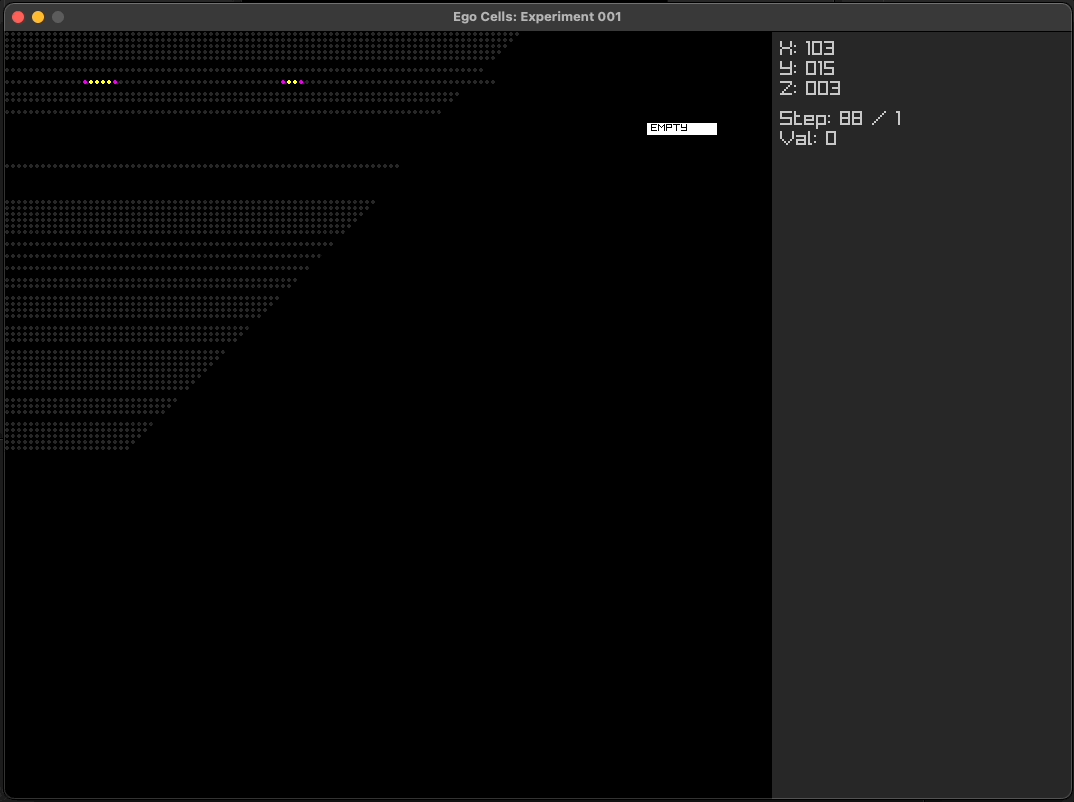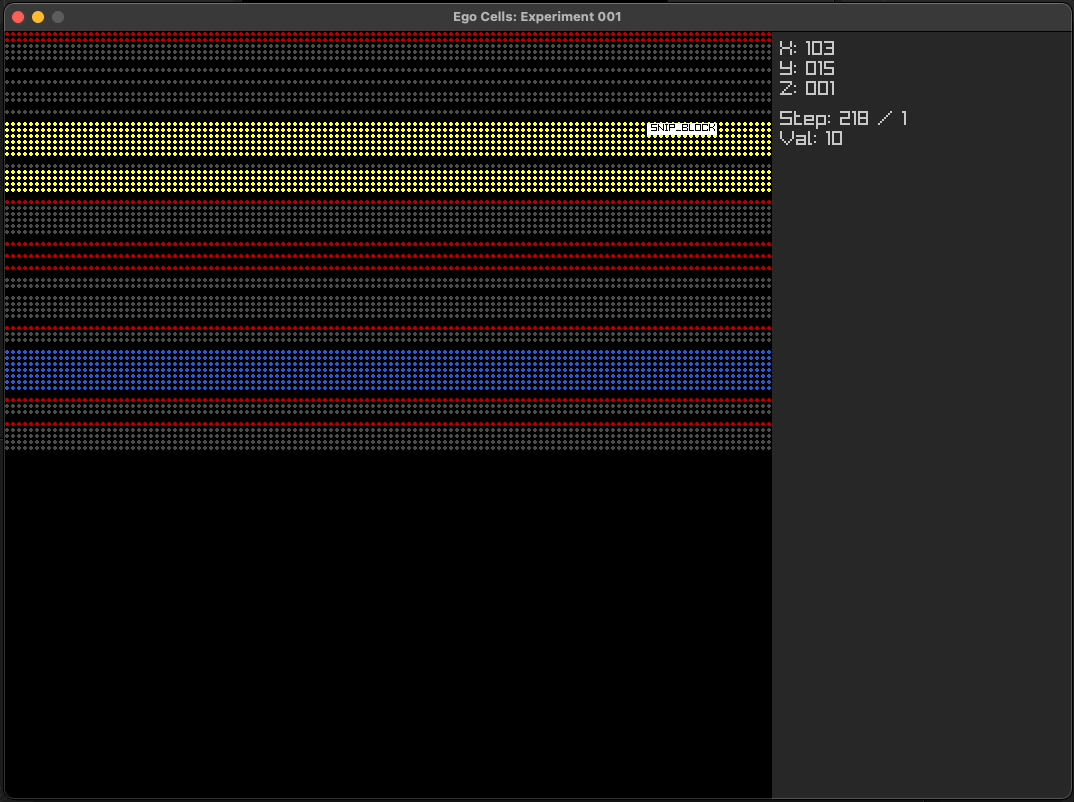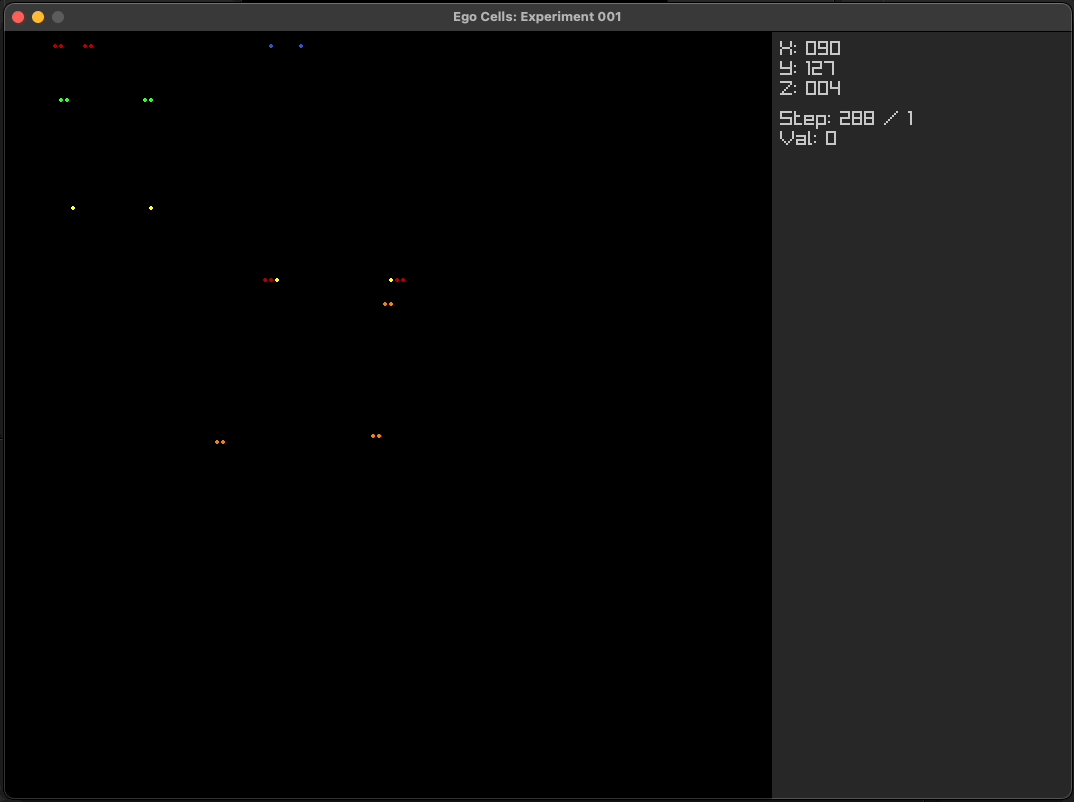
Using ego cell tissue as cortical columns
While I was working on this experiment I read 📑 "A Framework for Intelligence and Cortical Function Based on Grid Cells in the Neocortex" by Numenta. It proposes a theoretical framework to understand how the cortical columns in the neocortex might use possible grid cells to generate meaning in a space-oriented manner. I got hooked by this idea and started to think how this experiment could relate to that.
Even though this experiment only explores a part of what ego cells are meant to be, it was enough for me to start thinking about
Digital spatiotemporal substrate
properties of the digital space
- Text files, images, and computers in general
- Cells are wired accordingly to this nature
- Common spatial patterns are needed for goal setting
- borders / sense of orientation
A general layout for intelligence
- Active Modeling
- Attention Management
- Body Management
Prototyping pipeline
OpenCL and scalability
Final thoughts & further explorations
- UMMF (Universal Mind Map Format) — Model Translation
- Spatial Representation
- Relational Representation
- Associated Data Array
- Basic 2D grid world with simple life
Clip Notes & Vault Meta
Test tags
Clip Notes
- 📎 Correction Cells
- 📎 Analyzing the true essense of markdown
- 📎 Underestimating the parsing problem
- 📎 Memory and sequential patterns
- 📎 Spatial frameworks for modeling spatial systems
- 📎 Thinking in terms of simple programs
- 📎 Walls and limits — Are these place cells?
To achieve A, I did B. I observed C thus I conclude D.
Presenting initial assumption: Parsing MD with ego cells is possible by:
Having a grid of cells able to read their neighbors
Each cell needs access to their surrounding neighbors output
Where neighboring rules divide into abstraction layers
Character types
Token types
Block types
Each abstraction is defined by the neighboring rules that react to the layer below or above
Cells can arbitrarily only read 6 block radius each
In order to successfully detect all tokens, I tried using a radius of 3 blocks. But I noticed that there were some problems, so the radius maybe should not be defined explicitely.
Each abstraction layer should correspond to a z-level or 2D layer of cells forming a cubicical grid
Implementing neighboring rules for markdown
Setup development environment
Load markdown and have a buffer ready
Setup OpenCL
Adding visualizations
Writing neighboring rules
Trouble with finding the neighboring rules for higher abstractions
Trouble with detecting stable code blocks
Trouble with overriding rules
Re-approaching the problem
Analyzing the true essence of markdown
Orientation and sequence
Text types (code and none-code)
Nested text types (bold, non-bold, etc)
Wolfram's simple programs
Per-layer programs
Genetic Algorithms
Genes per simple program vs the whole
Concluding observations
Numenta's theory of a thousand brains
Cortical columns as a spatio-temporal mirroring substrate
Digital spacetime
Ego Cells as Cortical Columns
A possible framework for human-like intelligence: The three parts of a brain
Modeling substrate
Guiding system
Body maintenance system
Performance - GPUs, CPUs and FPGAs